Ductal carcinoma in situ (DCIS), a non-invasive form of breast cancer found in the milk ducts, is a precursor to invasive breast cancer, but until recently, its progression has remained enigmatic. This is partly because standard methods of preserving tissue—as formalin-fixed paraffin-embedded (FFPE) samples—have made single-cell genetic analysis difficult. Now, thanks to an FFPE-compatible DNA sequencing method developed by former Damon Runyon scientists Nicholas E. Navin, PhD, Runmin Wei, PhD, and their colleagues at MD Anderson Cancer Center, research has shed light on how DCIS evolves into invasive breast cancer.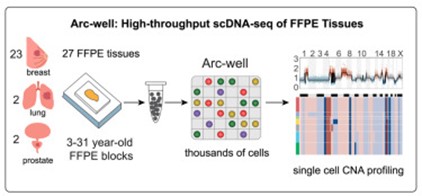
Using their sequencing method, known as Arc-well, the team profiled over 40,000 single cells, as well as frozen and FFPE samples from breast, lung, and prostate tumors stored for up to 31 years. This diverse sample set demonstrated the robustness of Arc-well across tissue types and storage durations.
The study’s most striking findings came from analyzing DCIS samples with matched breast cancer samples, recurring 2 to 16 years later, in a small group of patients. The results revealed that many primary DCIS cells had already undergone significant genomic alteration, and in some cases whole-genome doubling, prior to progression. As a result, the cell population in the DCIS samples was highly diverse.
Evolutionary analysis conducted by the researchers indicated that most DCIS cases in their cohort experienced an “evolutionary bottleneck,” meaning that, during the transition from DCIS to invasive cancer, the genetic diversity of cancer cells narrowed, leading to a few dominant subtypes. These dominant subtypes were found to possess genetic aberrations associated with cancer recurrence, providing valuable insights into potential therapeutic targets.
In addition to the study of breast cancer evolution, Arc-well has broad potential applications for studying existing archives of FFPE and frozen samples to elucidate the progression of other forms of cancer.
This research was published in Cell.







