Epidermal growth factor receptor (EGFR) is a protein on the surface of cells that receives signals telling the cell to grow. Mutations in the EGFR gene are known to drive a number of cancers, including non-small cell lung cancer. For patients with common EGFR mutations, known as “classical mutations,” EGFR inhibitor treatments are available and effective. But such targeted therapies have not been developed for patients with atypical mutations, often leaving chemotherapy as the only treatment option.
One challenge in designing clinical trials for new drugs is that patients are grouped according to where their mutation occurs in the EGFR gene, but in fact mutations occurring close together can result in significantly different mutant proteins, some more sensitive to drugs than others. This means drug developers are making decisions based on the wrong information—imagine a chef combining ingredients that sound similar rather than taste good together. A better system of classifying atypical mutations, one based on the mutant protein’s shape and drug sensitivity instead of the mutation’s location, is needed for clinical trials to produce meaningful results.
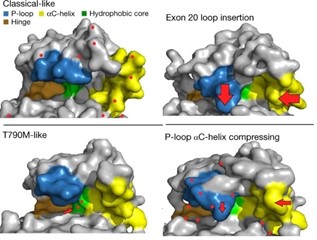
In their latest Nature paper, Damon Runyon-Lilly Clinical Investigator John Heymach, MD, PhD, and his team at MD Anderson Cancer Center provide just such a system. After collecting samples from nearly 17,000 patients with EGFR-mutant lung cancer, the team created 76 different mutant cell lines to target with available EGFR inhibitors. They observed which mutant cells succumbed to which drugs. From the results emerged four distinct subgroups of EGFR mutations, with classical mutations and those “classical-like” in shape ranking highest in sensitivity. Reviewing a database of lung cancer patients who had received targeted therapy, the researchers confirmed that, indeed, each patient’s outcome could be retrospectively predicted based on their mutation subgroup.
With this new classification system, clinicians can match patients with atypical EGFR mutations to the EGFR inhibitor that will be most effective for them, rather than resort to chemotherapy. The search for new drugs is also aided by these findings, as whole subgroups of mutations can be targeted at once. Dr. Heymach’s paper has implications for the broader world of cancer research as well, challenging the idea that adjacent mutations behave similarly and offering a more useful grouping principle for clinical trials and drug development.
Read more in Nature.







