The KRAS gene, responsible for encoding a protein that serves as an “on/off” switch for cell growth, is one of the most commonly mutated genes in cancer. The frequency and nature of its mutation differ across cancer types, however, with the highest occurrence of mutation found in cancers of the colorectum, pancreas, lung, and blood plasma.
We know from prior studies that specific mutant KRAS alleles correlate with different clinical outcomes. For example, pancreatic cancer patients who have the KRAS G12D allele tend to have a worse prognosis than those with different KRAS mutations; likewise, colorectal cancer patients with a KRAS G13D allele tend to respond better to certain therapies. While scientists have observed these correlations, the pathways that lead from specific alleles to varying cancer diagnoses, disease courses, and treatment responses are not well understood.
Seeking clarity that could only come from a vast data set, former Damon Runyon Fellow Kevin Haigis, PhD, and his lab at Dana-Farber Cancer Institute analyzed over thirteen thousand tumor samples from the four cancer types mentioned above, which most often carry KRAS mutations. Their findings not only show how different mutant KRAS alleles are distributed across cancers, but also indicate the other genetic events and biological processes that accompany these mutations: the sequence of events leading from mutation to clinical consequence.
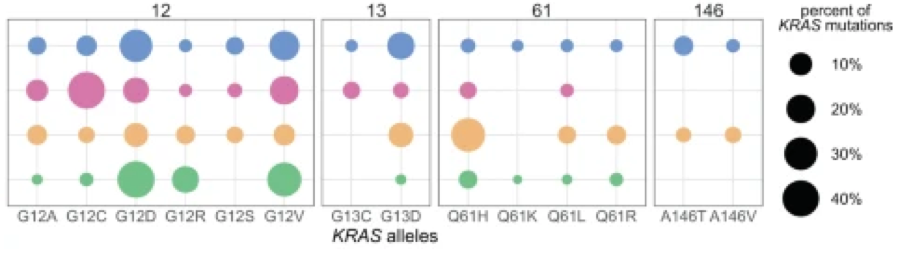
(blue = colorectum; pink = lung, gold = blood plasma; green = pancreas)
Slightly different KRAS alleles can result in very different clinical outcomes due to distinct “comutation networks”— groups of genes that mutate together and reinforce each other’s effects. Each mutant KRAS form has its own comutation network.
Interestingly, these allele-specific networks are not consistent between tissues, suggesting tissue-specific signals that also influence how the KRAS allele interacts with other genes. The discovery of both allele-specific and tissue-specific properties attending mutant KRAS forms highlights the complexity of KRAS-driven oncogenesis.
KRAS-driven cancers are notorious for thwarting precision medicine due to the variation in their signaling pathways. Thanks to Dr. Haigis’ lab, our sketch of the complex KRAS landscape has recently acquired more detail, revealing possible new targets against this “most potent oncogene.” From this understanding, too, more general principles may be derived about oncogenes with multiple sites of mutation.
This study was published in Nature last month.







